ssDNA (Single-Stranded DNA Synthesis)
ssDNA Synthesis service provides up to 10,000 nt of full sequence-verified fragments quickly and affordably. Our ssDNA Synthesis fragments are derived from clonally purified double-stranded DNA (dsDNA), producing the highest quality results possible.
Recent research in CRISPR/Cas9 technology has shown that the use of long ssDNA donor templates greatly enhances the efficiency of homology-directed repair (HDR) enabling researchers to optimize the process of efficiently generating transgenic animal models and cell lines. As the leader in complex gene sequences, you can trust GENEWIZ from Azenta Life Sciences for sequence-verified ssDNA synthesis for CRISPR gene knock-in, in-vitro transcription, and much more.
What is the purpose of ssDNA?
ssDNA, or single-stranded DNA, is a nucleotide sequence involved in DNA replication and DNA repair.
What is the difference between dsDNA and ssDNA?
dsDNA stands for double-stranded DNA, which is two complimentary DNA strands forming a helical shape bound by hydrogen bonds. ssDNA stands for single-stranded DNA, and has a lower stability compared to dsDNA. Due to its double-stranded nature and strong hydrogen bond linkage, dsDNA is resistant to formaldehyde (unlike ssDNA).
ssDNA Synthesis Applications
CRISPR Gene Knock-In Workflow
Double-stranded breaks are generated through CRISPR/Cas9 editing, then repaired by the endogenous cellular pathways of non-homologous end joining (NHEJ) and HDR. While the HDR pathway has consistently proven successful in copying genetic information via homologous recombination, insertion of exogenous genetic material is a challenge due to the inherent inefficiencies of HDR. Double-stranded DNA has historically been the template of choice for gene insertions, but recent research has shown the superiority of ssODNs as a donor template for HDR. Offering much higher efficiency to insert long sequences with shorter homology arms, ssDNA has become the preferred donor template for this process. GENEWIZ now offers the longest (up to 10,000 nt) ssDNA fragments on the market, allowing insertion of long sequences with high efficiency and reduced cellular toxicity or off-target integration compared to dsDNA donors.
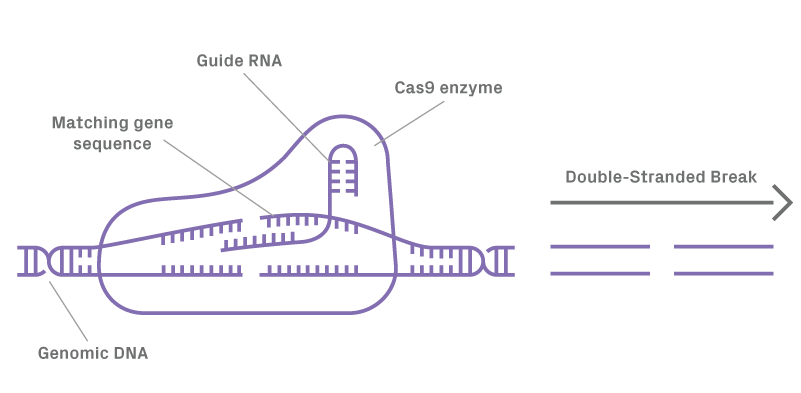
Figure 1.1. Guide RNA forms a complex with Cas9 directing enzyme to cleave target DNA resulting in a double-stranded break (DSB).
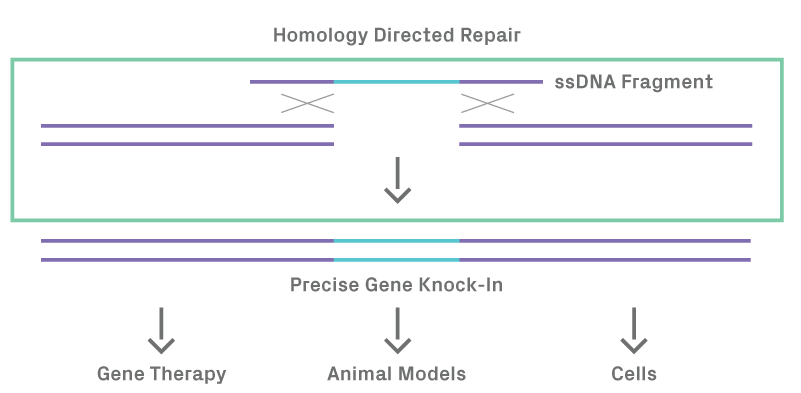
Figure 1.2. Homology directed repair after DSB in the presence of a ssDNA donor template results in precise gene knock-in.
ssDNA Advantages for CRISPR Gene Editing
Fastest and Longest ssDNA Available
| Length | Available Yield | Turnaround Time |
| 151-500 nt | 2, 3, 6, 10, 20, or 40 µg | Starting at 10 Business Days |
| 501 – 2,000 nt | 3, 6, 10, 20, or 40 µg | Starting at 15 Business Days |
| 2,001 – 8,000 nt | Starting at 20 Business Days |
Please note: 50% off complementary sequence for every order. Custom discount available for customer-supplied dsDNA.
Email gs@azenta.com to submit custom inquiries for orders >8,000 nt.
Single-Stranded DNA Features & Benefits
ssDNA Deliverables
- Sequence verified
- 2, 3, 6, 10, 20, or 40 µg lyophilized DNA
- Certificate of Analysis (COA) including gel image, sequence trace data with alignment, and sequence files
ssDNA Quality Control
- Sequence confirmation via Sanger sequencing
- Size verification by gel electrophoresis
- S1 nuclease digestion test
Webinar | Understanding biological mechanisms to better predict the evolution of antibiotic resistance
The rapid and wide-spread evolution of antibiotic resistance is threatening global health. In this webinar, presented by Dr. Mato Lagator, Wellcome Trust, Royal Society Sir Henry Dale Fellow, University of Manchester, you will learn about different approaches to study evolutionary processes that underpin the emergence of resistance. Special focus is given on how these methods can be utilized to improve drug development and longevity.
Watch Now

