Primer Walking
Primer Walking utilizes multiple primers in a series to sequence entire plasmids or PCR products. GENEWIZ Primer Walking service options include plasmid sequence confirmation for already known sequences and plasmid sequence discovery for unknown sequences. For AAV vectors, we offer primer walking plus our new, proprietary AAV-ITR Sequencing. Trust GENEWIZ from Azenta to manage your project from primer design to variance analysis and report generation.
PLASMID SEQUENCE CONFIRMATION service
GENEWIZ primer walking sequence confirmation service offers peace of mind and helps you perform plasmid identity verification thus avoiding costly complications with downstream applications. Estimated turnaround time is 3-5 business days after primer design. We also offer GLP level plasmid confirmatory sequencing.
With our plasmid confirmation service you can:
• Confirm the sequence of your cloned products/genes of interest
• Sequence your entire plasmid, including difficult-to-amplify regions
• Examine failed amplifications and incorrect restriction digest patterns
• Sequence full length AAV plasmids and hard to sequence ITR regions

1. Submit Samples
2. Primer Design*
3. Sanger Sequencing

4. Consensus Sequence Assembly

5. Data Report
*Please note, a reference sequence is required for primer design.
PLASMID SEQUENCE DISCOVERY service
GENEWIZ primer walking sequence discovery service maps your unknown plasmid or PCR product for use in downstream applications. Subsequent primers will be designed as sequences are generated. Estimated turnaround time is 3-5 days per 1kb.
With our plasmid discovery service you can:
• Create a plasmid map for your undetermined plasmids
• Confirm the sequence of your mystery constructs
• Identify unknown regions of your plasmids
• Sequence full length AAV plasmids and hard to sequence ITR regions



*As a starting point, an antibiotic resistance gene, a known promoter (M13, T7), or another known sequence will be required for the initial primer design.
AAV-ITR Sequencing CAPABILITY
GENEWIZ has developed a proprietary Sanger sequencing method specifically designed to sequence the inverted terminal repeat (ITR) hairpin regions of adeno-associated virus (AAV) plasmids. Our proprietary protocol prevents the abrupt reduction in sequencing signal at the start of the ITR hairpin and reads through the full length of the ITR region.
AAV-ITR sequencing can be incorporated into either our sequence discovery or sequence confirmation services.
ITR sequencing using standard protocol
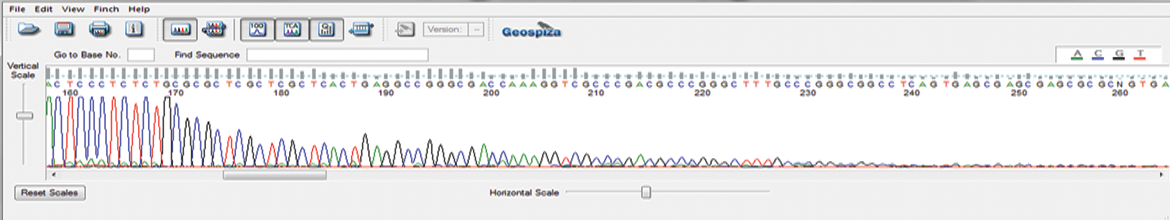
ITR Sequencing using GENEWIZ’s proprietary protocol
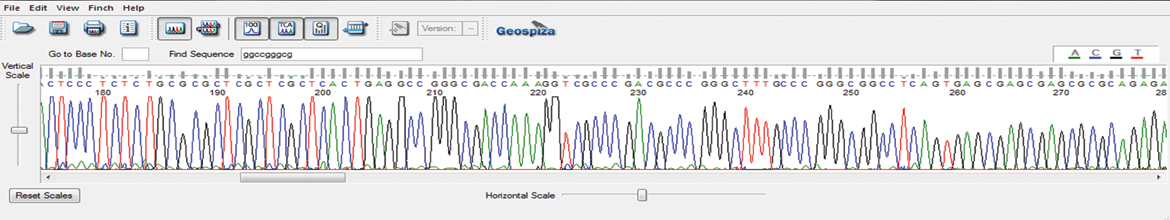
Primer Walking Features & Benefits
Technical Resources

Tech Note: Reading Through the Inverted Terminal Repeats (ITRs) of Adeno-Associated Virus (AAV)
GENEWIZ has developed a high-quality, direct Sanger sequencing method that reads through both intact and commonly mutated inverted terminal repeat (ITR) regions of adeno-associated virus (AAV). This method is an effective tool for assessing the integrity of ITRs in AAV plasmids.
Blog | Optimizing AAV Plasmid Preparation and ITR Sequencing
The adeno-associated virus (AAV) is a powerful vehicle for gene therapy; however, working with AAV vectors can be challenging. Each construct contains two inverted terminal repeat (ITR) sequences, usually 145 bp in length, that flank the gene of interest. Learn about the challenges of preparing and validating plasmid DNA for AAV vectors containing ITRs and an optimized approach to these processes.

Blog | Troubleshooting DNA Templates with Sanger Sequencing
Sanger sequencing is highly effective for testing small targeted genomic regions and for validating results from NGS. However, as with all sequencing technologies, there are limitations and needs for troubleshooting. Read on to learn about the three basic troubleshooting steps we recommend to start with.
