Sanger sequencing FAQs
Customer Accounts | DNA Sequencing Sample Submission & Primer | DNA Sequencing Results | Troubleshooting | Regulatory DNA Sequencing Services | Sanger-EZ | Low Concentration Plasmid
CUSTOMER ACCOUNTS
If this is the first time you are placing an order with GENEWIZ from Azenta, please note that the GENEWIZ list price may be displayed for your account. Institution-specific pricing will be available once your account is appropriately linked to your institution. If you have any questions about the price of your order, please contact dnaseq@azenta.com or your Sales Executive.
DNA SEQUENCING SAMPLE SUBMISSION & PRIMER
Choose “Pre-Mixed” if you have adjusted your sample concentration following our guidelines and have already added your own sequencing primers.
Choose “Pre-Defined” if you have adjusted your sample concentration following our guidelines but are supplying your primer in a separate tube for GENEWIZ from Azenta to add (small additional charge may apply) or are requesting a GENEWIZ free universal primers.
Choose “Custom” if you want GENEWIZ from Azenta to adjust your sample concentration and add your primers (additional charge may apply).
Choose “Custom” if you are requesting sequencing from bacterial colonies, glycerol stock, or phage; as well as if you are submitting un-purified PCR templates.
1.) Closely follow the DNA Sequencing Sample Submission Guidelines. A sub-optimal DNA-to-primer ratio is often the culprit for poor quality results.
2.) Make sure that your primer can adequately bind to your template. Insufficient primer binding can lead to disappointing results. Sequencing primers should be about 18-24 bases in length with a Tm of 56-60 degrees. The GC content should be about 45-55%, which is generally needed in order to get an 18-24 base primer in the Tm of 56-60.
3.) Avoid inhibiting contaminants. Salts, EDTA, alcohol, protein, RNA, detergents, cesium and phenol are some of the most common contaminants that can cause poor quality or failed reactions.
4.) Be sure to choose the correct protocol. Some templates (hairpins, GC-rich, etc.) benefit from a special protocol to ensure a good sequencing reaction. We offer alternative protocols at an additional cost that are highly effective in sequencing through difficult regions. These protocols can be requested from the “Special Requests” drop down menu on your online submission form.
We provide these free universal primers. If you are uncertain which of our universal primers bind to your construct, check out our universal primer selection tool. Simply copy and paste your FASTA sequence into the box, and the tool will display which primers will bind to your template. You can find the universal primer selection tool here.
View our Sample Storage Policy for more details.
- Plasmids and PCR (purified and unpurified) samples are stored for 5 business days (1 calendar week) from date of receipt. Primers received along with the plasmids and PCR samples are stored for the same duration.
- Bacterial colonies, glycerol stocks, and phage are stored for 10 business days (2 calendar weeks) from date of receipt. Primers received along with these samples are stored for the same duration.
- Oligo Retain for Sanger (oligos synthesized by GENEWIZ) are stored for 1 year from date of synthesis.
Nunc (through Fisher): 96-Well PCR Plates- 12-565-536, 12-566-133, or 12-566-134, Caps- 12-565-810 or 12-575-123
Nunc (through VWR): 96-Well PCR Plates- 73520-694, 83009-280, or 73520-696, Caps – 73521-364 or 73521-066
USA scientific- Plates- 1402-9400, 1402-9600, 1402-9700 with caps- 1400-0800 or 1400-3800
ABI- Plates 4306737, Caps 4323032
DNA SEQUENCING RESULTS
The .ab1 file is your chromatogram, and requires chromatogram viewing software to view these files. Please visit https://www.genewiz.com/public/resources/tools-for-viewing-sequencing-data (or click on the “Tools for Viewing Sequencing Data” link on the menu) for a number of free software programs that are available for viewing trace or chromatogram files.
TROUBLESHOOTING
REGULATORY DNA SEQUENCING SERVICES
SANGER-EZ
You will receive an email with a link to view the results when available.
IMPORTANT: The barcode number of the tube will be used as the DNA/Sample Name when viewing the results. Please make sure to record this number prior to submission.
2. Sanger-EZ tubes can be submitted to either a GENEWIZ dropbox or shipped to a GENEWIZ facility. IMPORTANT: if submitting to a “By-Request” GENEWIZ dropbox, a pickup request must be placed to ensure sample pickup by our courier. Pick-up requests are made within your GENEWIZ account under “DROP-OFF AND DELIVERY BOXES”.
3. Ensure to note the barcode number for each reaction. This number will act as the sample name to view results after processing.
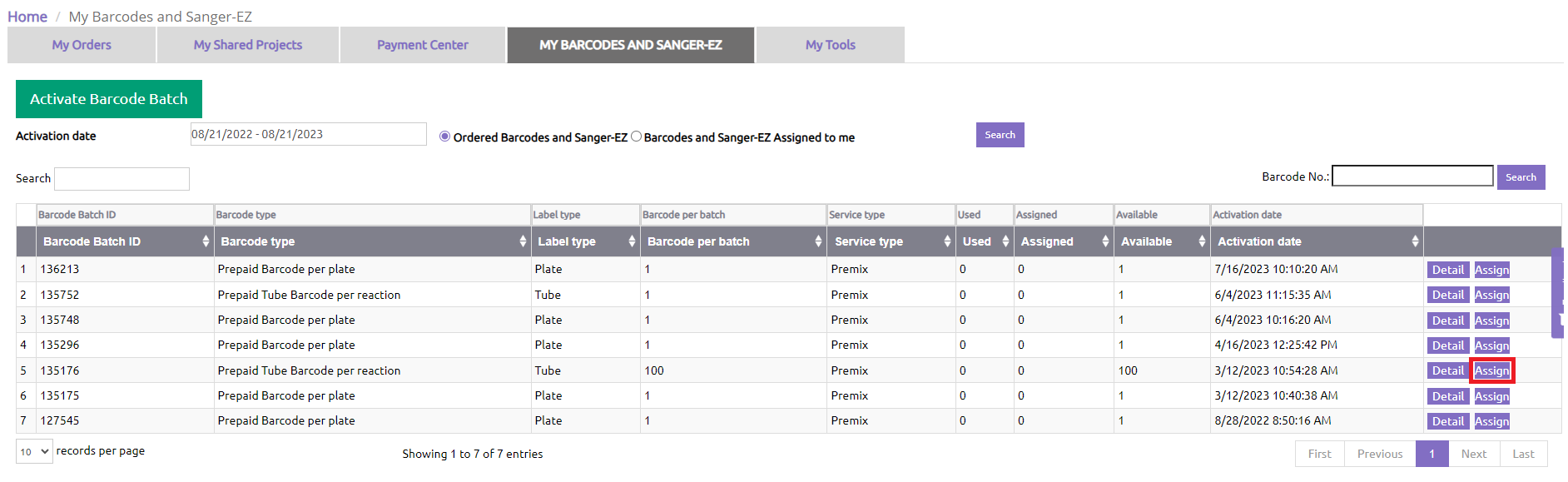
Find the Sanger-EZ batch you would like to share and click “Assign”.
Select 1 or more colleagues and follow instructions to share Sanger-EZ tubes.