ATAC-Seq
ATAC-Seq (assay for transposase-accessible chromatin sequencing) identifies open chromatin regions and efficiently labels them for high-throughput sequencing. This technique has a higher signal-to-noise ratio and lower input requirements than similar approaches.
Our ATAC-Seq service, from library preparation to data analysis, provides an optimized approach for nuclei extraction from cryopreserved cells to ensure detailed insight into genome-wide chromatin accessibility. If you want to understand gene expression regulation at the individual-cell level in varied states and cellular processes, we also offer single-cell ATAC-Seq (scATAC-Seq).
ATAC-Seq Workflow
1. Sample Preparation
Isolate and freeze cells with protocol provided by GENEWIZ
2. Dead Cell Removal
If low viability, live cells are enriched using a magnetic bead-based approach
3. Nucleic Extraction & Library Preparation
Optimized ATAC assay produces robust signal from low inputs
4. Sequencing
Guaranteed data quality exceeds Illumina® benchmarks
5. Bioinformatics
Alignment, peak calling, and multiomic data integration
ATAC-Seq Features & Benefits
ATAC-Seq Overview
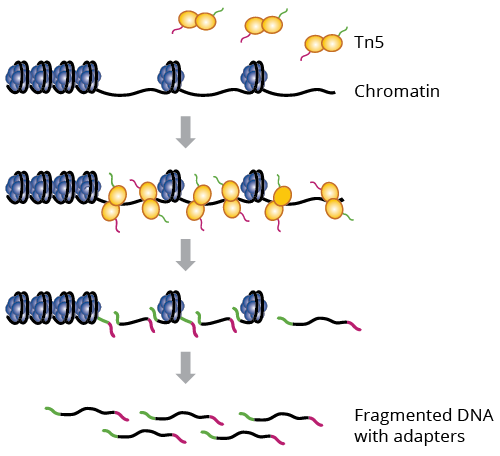
Assay for transposase-accessible chromatin sequencing (ATAC-Seq) employs a hyperactive form of Tn5 transposase to identify regions of open chromatin, which are important for global epigenetic control of gene expression. Tn5 simultaneously cleaves and adds adapters to nucleosome-free regions of DNA, priming them for sequencing.
NGS PLATFORMS
For information on our NGS platforms as well as recommended configurations of your projects, please visit the NGS Platforms page. GENEWIZ from Azenta does not guarantee data output or quality for sequencing-only projects.