Single-Cell ATAC-Seq (scATAC-Seq)
Single-cell ATAC-Seq (scATAC-Seq) is used to investigate chromatin accessibility in hundreds to thousands of individual nuclei from a single sample. Unlike bulk ATAC-Seq (assay for transposase-accessible chromatin sequencing), which obscures heterogeneous cell populations in a single transposition reaction, scATAC-Seq utilizes microfluidics and molecular barcoding to identify specific chromatin signatures in individual cells. This approach helps to discover regulatory patterns and offers a deeper understanding of gene regulation.
Our scATAC-Seq service utilizes the latest technology on the market: 10x Genomics®, Illumina®, and PacBio® platforms. As an early adopter of these platforms, our optimized workflows — including pre-submission cryopreservation and post-submission dead cell removal — maximize project flexibility, speed, and data accuracy while eliminating costly overhead and ramp-up time to expedite your research.
For a single-cell multiomic approach, our scATAC-Seq and single-cell RNA-Seq (scRNA-Seq) services can be combined in an integrated data analysis framework to uncover novel biological features.
WHAT IS SINGLE-CELL ATAC-SEQ?
Assay for transposase-accessible chromatin sequencing (ATAC-Seq) identifies regions of open chromatin and efficiently labels them for high-throughput sequencing. Single-cell ATAC-Seq allows researchers to identify these regions within individual cells.
WHAT DOES SINGLE-CELL ATAC-SEQ TELL YOU?
Single-cell ATAC-Seq identifies chromatin accessibility in the epigenome by profiling thousands of single cells in parallel, enabling discovery of how chromatin structure and DNA-binding proteins regulate gene expression in varying states and cellular processes.
HOW DOES SINGLE-CELL ATAC-SEQ WORK?
Following dead cell removal using a magnetic bead-based approach to enrich live cells, cells and barcoded beads are isolated in droplets using the 10x Genomics Chromium X and then tagmented. Unique barcodes for each individual cell are added to DNA prior to library preparation. Once library preparation is completed, barcoded libraries are pooled and sequenced on the Illumina NovaSeq for high-throughput characterization.
scATAC-SEQ WORKFLOW
-

STEP 1.
DEAD CELL REMOVALIf viability and cell count are low, dead cells are removed using a magnetic bead-based approach to enrich live cells.
-

STEP 2.
CELL PARTITIONINGCells and barcoded beads are isolated in droplets using the 10x Genomics Chromium and tagmented.
-

STEP 3.
LIBRARY PREPARATIONUnique barcodes for each individual cell are added to DNA prior to library preparation.
-

STEP 4.
SEQUENCINGBarcoded libraries are pooled and sequenced on the Illumina NovaSeq for high-throughput characterization.
-
STEP 5.
BIOINFORMATICS DATA ANALYSISVisualize results through interactive software and receive custom analysis.
scATAC-SEQ: EPIGENOMIC INSIGHTS AT THE CELLULAR LEVEL
-
Cell Cluster and Classification

Identify and classify diverse cell types in heterogenous cell populations.
-
Trajectory Inference
Discover states for lineage and development tracing to understand mechanisms of cell fate.
-
Regulatory Variation
Detect altered gene regulation and/or novel enhancers in rare cells and sub-populations.
-
Chromatin Accessibility
Reveal subpopulation-specific regulation, enabling prediction of cell type-specific, cis-regulatory networks.
FEATURES & BENEFITS
-
Superior Data Quality
Exceeding manufacturer’s benchmarks -
Real-Time Project Updates
Through our online system -
Ph.D.-Level Support
At every step
-
Proprietary cell freezing method maintains cell viability during storage and transit.
-
Dead cell removal protocol improves data quality, especially for samples with suboptimal viability.
-
Early adopter of the 10x Genomics Chromium with optimized workflows that maximize project flexibility, speed, and data accuracy.
-
Highest-throughput sequencing platforms, including the Illumina NovaSeq, provide cost-effective sequencing solutions.
-
Flexible starting materials accepted including cell pellets with as few as 10,000 viable cells and frozen tissue; isolated nuclei are not required.
-
Samples accepted at different stages of the 10x workflow including cells, Gel Beads in Emulsion (GEMs), or 10x barcoded cDNA libraries for maximum workflow flexibility.
SINGLE-CELL MULTIOME
Integrate scRNA-Seq and scATAC-Seq data analysis frameworks to quantify how chromatin structure influences the regulation of gene expression. Download our poster about single-cell multi-omic profiling of cryopreserved tissue to learn more.
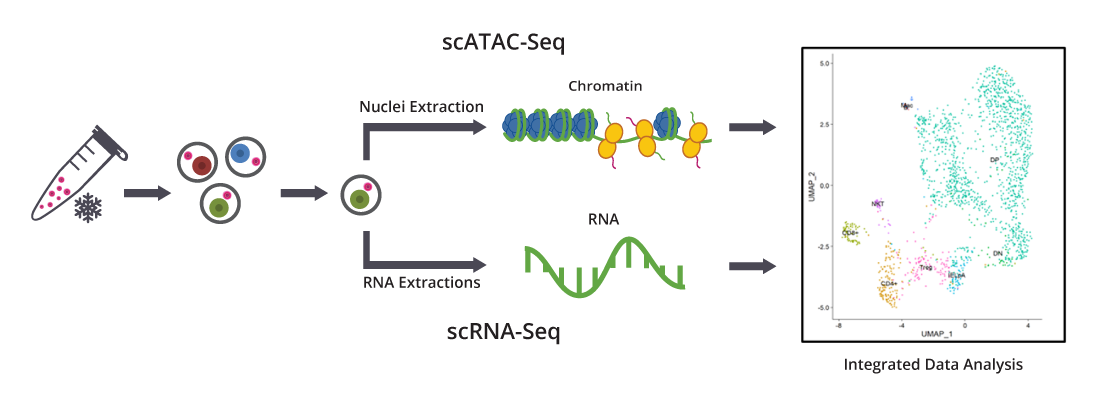
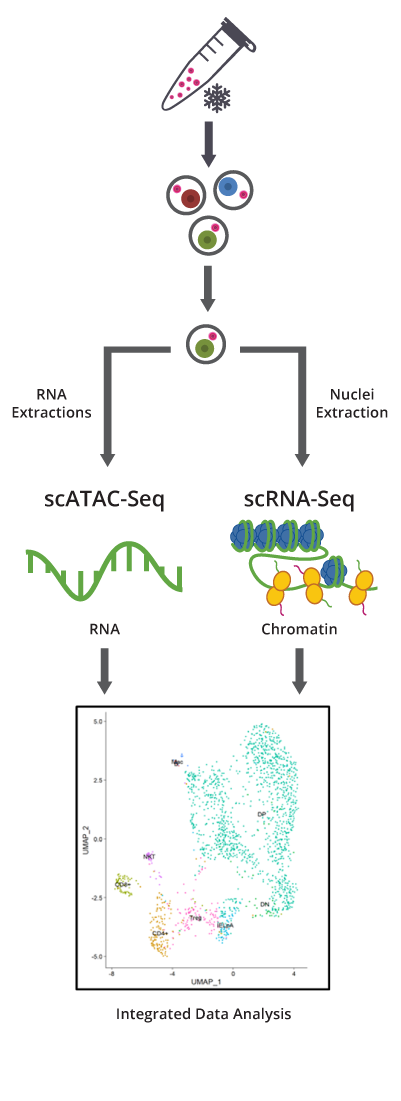
Figure: Frozen cells undergo our proprietary dead cell removal protocol. Viable cells are sorted, followed by nuclei and RNA extraction, and are subjected to scATAC-Seq and scRNA-Seq, respectively. Integrative data analysis of chromatin accessibility and gene expression for the same cell types can be used for confirming cell identity annotation by UMAP projections. The gene activity matrix derived from counts in gene body and promoter regions determines putative cell types.
SINGLE-CELL MULTI-OMICS BENEFITS
-
Streamlined experimental design with sample splitting to reduce variability between assays.
-
Integrated data analysis provides greater insight into gene regulation and enables more complete sample characterization.
-
Reduced cost and faster results from combined workflows compared to separate analyses.
-
Customized and extensive bioinformatic solutions with interactive, hands-on analysis and publication-ready images.
SINGLE-CELL ATAC-SEQ TECHNICAL RESOURCES
-

Poster: Highly Multiplexed Single-Cell Transcriptome and Epigenome Profiling of Cryopreserved Tissue Enables Multi-omic Tissue Characterization in Clinical Settings
Single-cell analysis typically relies upon immediate processing of fresh tissue, presenting limitations for translational and clinical applications. Learn how our robust cryopreservation workflow for downstream processing enabled high-quality, single-cell transcriptomic and epigenomic analysis of translational and clinical samples by coupling scATAC-Seq and scRNA-Seq.
-

Tech Note: Add More Life to Your Data: Optimizing Single-Cell RNA-Seq with Dead Cell Removal
Obtaining samples with high cell viability can be difficult for many experiments but is necessary for success on the 10x Genomics Chromium platform. This tech note describes how GENEWIZ scientists used optimized single-cell workflows, including dead cell removal, to overcome low viability and generate high-quality sequencing data.
-

Top 3 Factors to Consider Before Starting a Single-Cell Sequencing Project
With several commercial platforms available, high-throughput single-cell sequencing is now more accessible than ever. In this article, we’ll touch upon the 3 most important factors to consider before embarking on your single-cell project to ensure you select the right NGS approach for your research.
NGS PLATFORMS
For information on our NGS platforms as well as recommended configurations of your projects, please visit the NGS Platforms page. GENEWIZ from Azenta does not guarantee data output or quality for sequencing-only projects.





